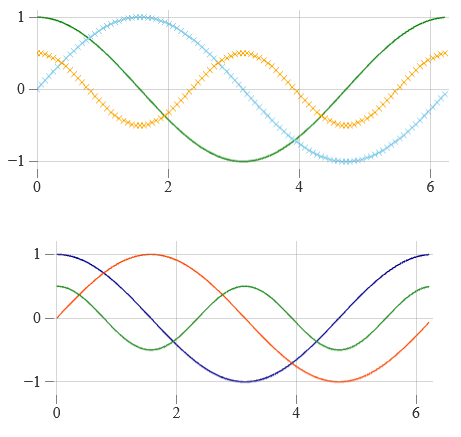
High-level components (heatmap, line, etc.) for use in existing React apps
🖼️ GPU rendering with WebGL (via three.js and react-three-fiber)
Low-level components (canvas, zoom, etc.) for advanced use cases


Loïc Huder
European Synchrotron Radiation Facility, Grenoble, France

 This project has received funding from the European Union’s
Horizon 2020 research and innovation programme under grant
agreement No. 823852
This project has received funding from the European Union’s
Horizon 2020 research and innovation programme under grant
agreement No. 823852
HDF5 (with NeXus) is now the standard format for acquired data at ESRF
👉 Need for a software to efficiently visualise and browse HDF5 file contents in the web and to be flexible for easy integration
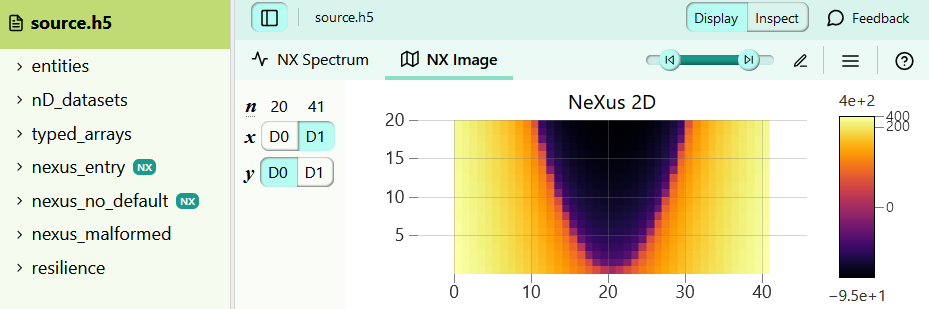
 @h5web/lib, the data visualisation library
@h5web/lib, the data visualisation library
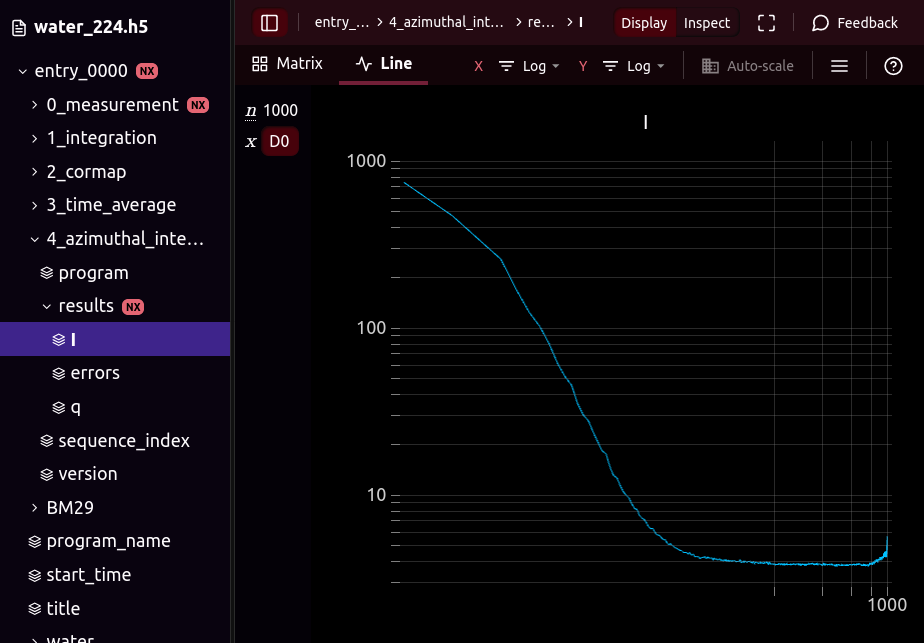
 @h5web/app, the viewer
@h5web/app, the viewer
High-level components (heatmap, line, etc.) for use in existing React apps
🖼️ GPU rendering with WebGL (via three.js and react-three-fiber)
Low-level components (canvas, zoom, etc.) for advanced use cases
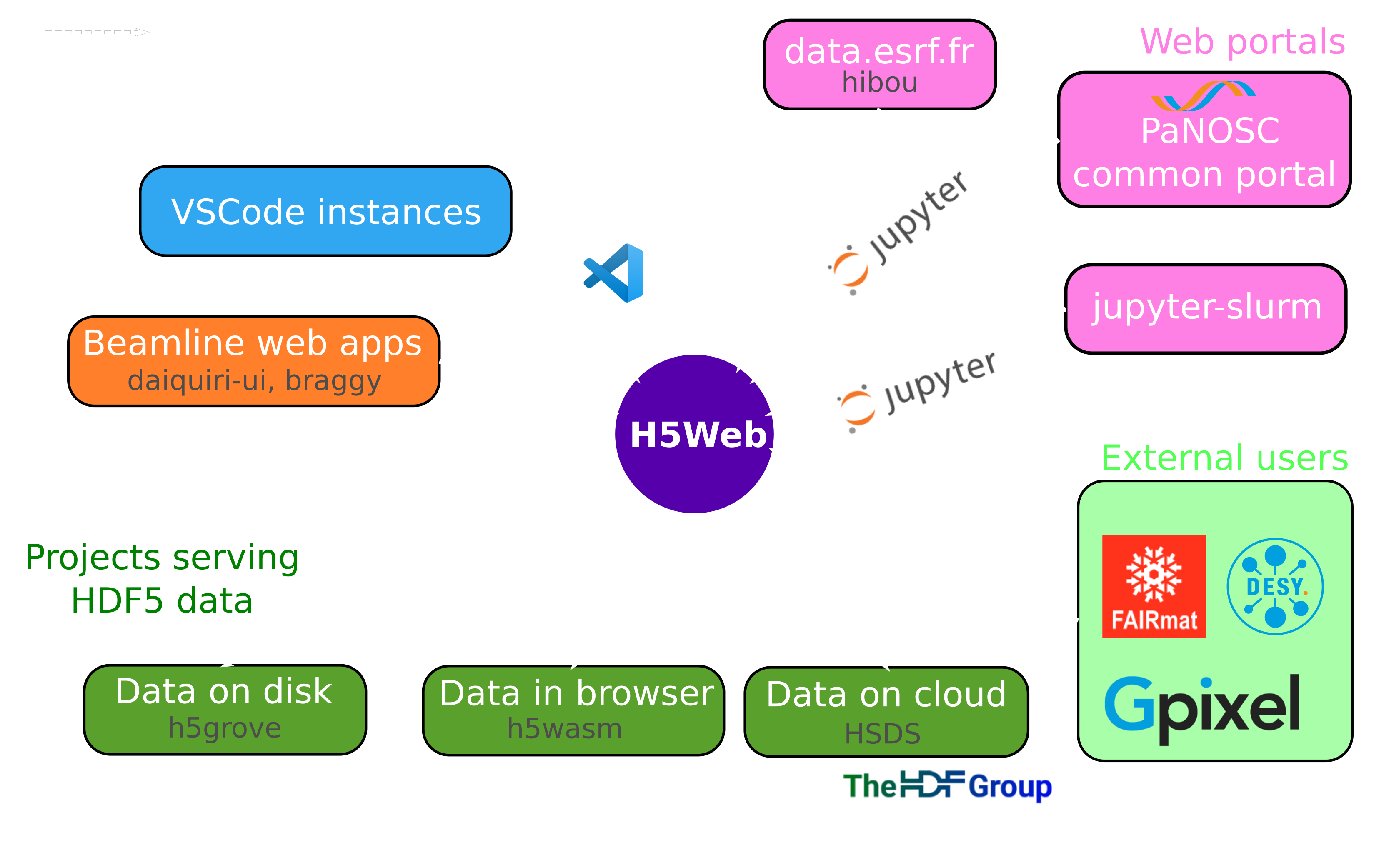
you can get involved!
🤔 You tried to install the JupyterLab/VSCode extension to browse your files and want to tell us how it went?
🤔 You have a usecase for H5Web that you want to discuss?
Questions ?
 This project has received funding from the European Union’s
Horizon 2020 research and innovation programme under grant
agreement No. 823852
This project has received funding from the European Union’s
Horizon 2020 research and innovation programme under grant
agreement No. 823852

Challenges
How H5Grove helps